Re: Parallel Seq Scan vs kernel read ahead
| От | David Rowley |
|---|---|
| Тема | Re: Parallel Seq Scan vs kernel read ahead |
| Дата | |
| Msg-id | CAApHDvqZbhmcuNWtuOd7sFynF=b02fVNtM60amVTgSmVkKQ08g@mail.gmail.com обсуждение исходный текст |
| Ответ на | Re: Parallel Seq Scan vs kernel read ahead (David Rowley <dgrowleyml@gmail.com>) |
| Ответы |
Re: Parallel Seq Scan vs kernel read ahead
(Robert Haas <robertmhaas@gmail.com>)
|
| Список | pgsql-hackers |
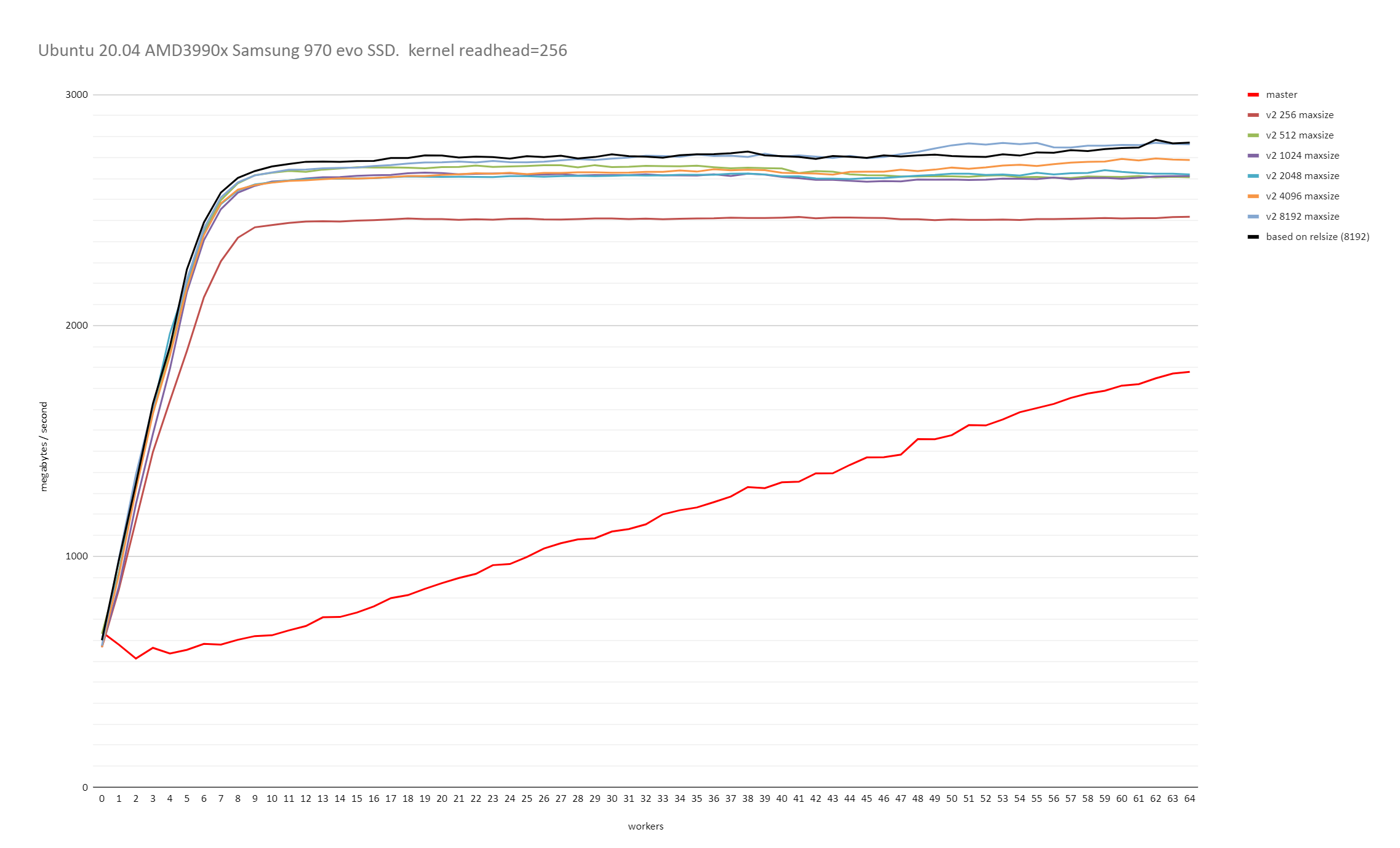
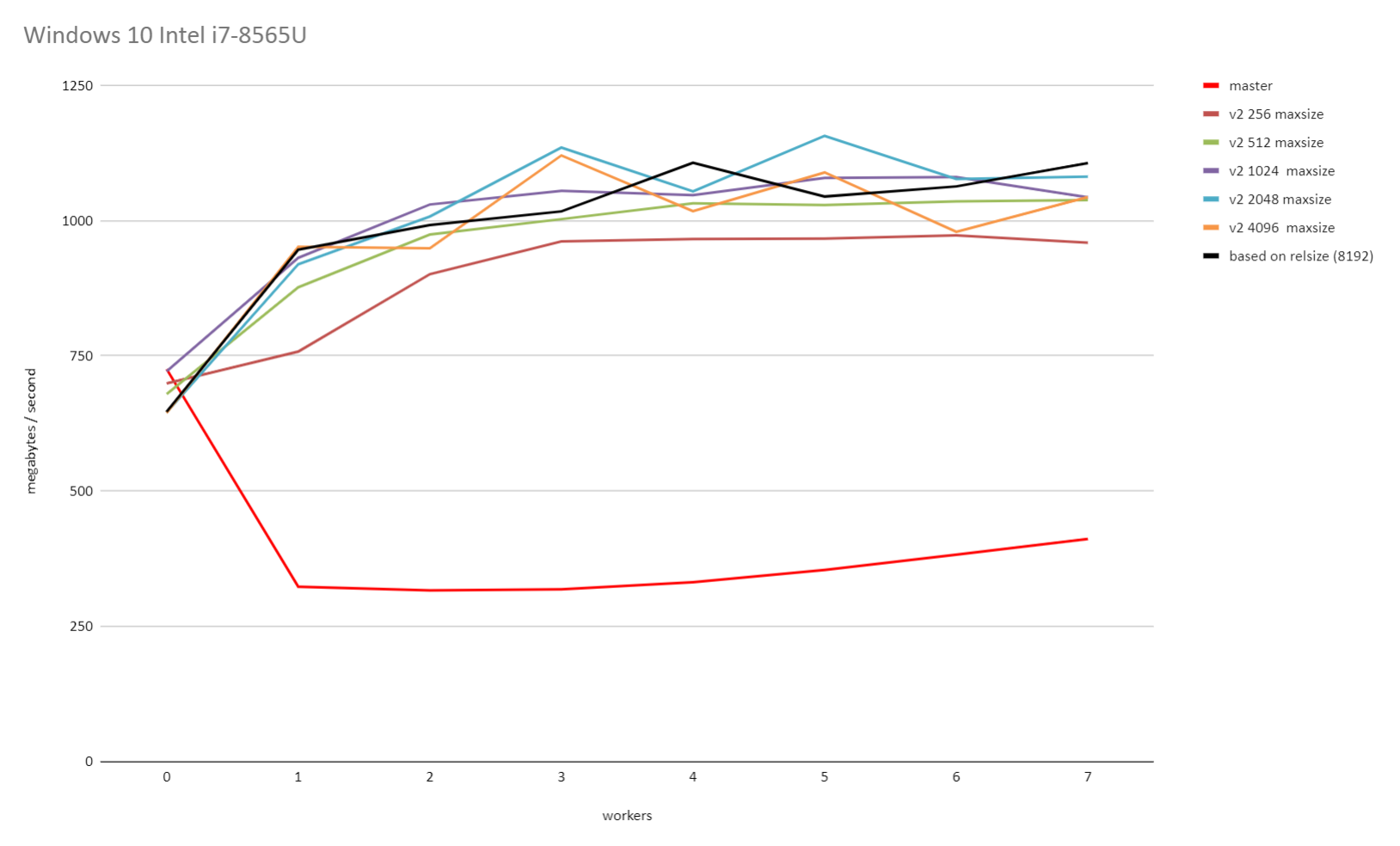
On Tue, 23 Jun 2020 at 10:50, David Rowley <dgrowleyml@gmail.com> wrote: > > On Tue, 23 Jun 2020 at 09:52, Thomas Munro <thomas.munro@gmail.com> wrote: > > > > On Fri, Jun 19, 2020 at 2:10 PM David Rowley <dgrowleyml@gmail.com> wrote: > > > Here's a patch which caps the maximum chunk size to 131072. If > > > someone doubles the page size then that'll be 2GB instead of 1GB. I'm > > > not personally worried about that. > > > > I wonder how this interacts with the sync scan feature. It has a > > conflicting goal... > > Of course, syncscan relies on subsequent scanners finding buffers > cached, either in (ideally) shared buffers or the kernel cache. The > scans need to be roughly synchronised for that to work. If we go and > make the chunk size too big, then that'll reduce the chances useful > buffers being found by subsequent scans. It sounds like a good reason > to try and find the smallest chunk size that allows readahead to work > well. The benchmarks I did on Windows certainly show that there are > diminishing returns when the chunk size gets larger, so capping it at > some number of megabytes would probably be a good idea. It would just > take a bit of work to figure out how many megabytes that should be. > Likely it's going to depend on the size of shared buffers and how much > memory the machine has got, but also what other work is going on that > might be evicting buffers at the same time. Perhaps something in the > range of 2-16MB would be ok. I can do some tests with that and see if > I can get the same performance as with the larger chunks. I did some further benchmarking on both Windows 10 and on Linux with the 5.4.0-37 kernel running on Ubuntu 20.04. I started by reducing PARALLEL_SEQSCAN_MAX_CHUNK_SIZE down to 256 and ran the test multiple times, each time doubling the PARALLEL_SEQSCAN_MAX_CHUNK_SIZE. On the Linux test, I used the standard kernel readhead of 128kB. Thomas and I discovered earlier that increasing that increases the throughput all round. These tests were done with the PARALLEL_SEQSCAN_NCHUNKS as 2048, which means with the 100GB table I used for testing, the uncapped chunk size of 8192 blocks would be selected (aka 16MB). The performance is quite a bit slower when the chunk size is capped to 256 blocks and it does increase again with larger maximum chunk sizes, but the returns do get smaller and smaller with each doubling of PARALLEL_SEQSCAN_MAX_CHUNK_SIZE. Uncapped, or 8192 did give the best performance on both Windows and Linux. I didn't test with anything higher than that. So, based on these results, it seems 16MBs is not a bad value to cap the chunk size at. If that turns out to be true for other tests too, then likely 16MB is not too unrealistic a value to cap the size of the block chunks to. Please see the attached v2_on_linux.png and v2_on_windows.png for the results of that. I also performed another test to see how the performance looks with both synchronize_seqscans on and off. To test this I decided that a 100GB table on a 64GB RAM machine was just not larger enough, so I increased the table size to 800GB. I set parallel_workers for the relation to 10 and ran: drowley@amd3990x:~$ cat bench.sql select * from t where a < 0; pgbench -n -f bench.sql -T 10800 -P 600 -c 6 -j 6 postgres (This query returns 0 rows). So each query had 11 backends (including the main process) and there were 6 of those running concurrently. i.e 66 backends busy working on the problem in total. The results of that were: Auto chunk size selection without any cap (for an 800GB table that's 65536 blocks) synchronize_seqscans = on: latency average = 372738.134 ms (2197.7 MB/s) <-- bad synchronize_seqscans = off: latency average = 320204.028 ms (2558.3 MB/s) So here it seems that synchronize_seqscans = on slows things down. Trying again after capping the number of blocks per chunk to 8192: synchronize_seqscans = on: latency average = 321969.172 ms (2544.3 MB/s) synchronize_seqscans = off: latency average = 321389.523 ms (2548.9 MB/s) So the performance there is about the same. I was surprised to see that synchronize_seqscans = off didn't slow down the performance by about 6x. So I tested to see what master does, and: synchronize_seqscans = on: latency average = 1070226.162 ms (765.4MB/s) synchronize_seqscans = off: latency average = 1085846.859 ms (754.4MB/s) It does pretty poorly in both cases. The full results of that test are in the attached 800gb_table_synchronize_seqscans_test.txt file. In summary, based on these tests, I don't think we're making anything worse in regards to synchronize_seqscans if we cap the maximum number of blocks to allocate to each worker at once to 8192. Perhaps there's some argument for using something smaller than that for servers with very little RAM, but I don't personally think so as it still depends on the table size and It's hard to imagine tables in the hundreds of GBs on servers that struggle with chunk allocations of 16MB. The table needs to be at least ~70GB to get a 8192 chunk size with the current v2 patch settings. David
Вложения
В списке pgsql-hackers по дате отправления:
Следующее
От: Michael PaquierДата:
Сообщение: Re: Update InsertPgAttributeTuple comment to match new signature